Segmentation in Transmitted-Light Images
21 December 2021
This is a novel cell segmentation method that tackles the difficult case in which neither a cell marker nor phase-contrast are available.
|

|
|
Context
When imaging fluorescent signals, phase-contrast is often not used because it reduces the fluorescence signal intensity. The easiest option to segment cells (in terms of image analysis) is then to use a fluorescent marker, but this can have a high experimental cost. An option is then to use defocused transmitted-light images to segment cells. Those images are much less contrasted than the two options mentioned above, and thus segmentation is much more difficult especially with dense cell colonies.
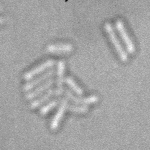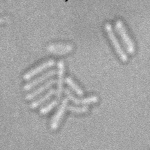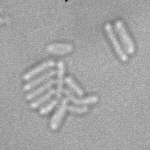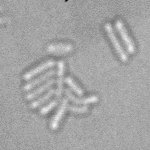
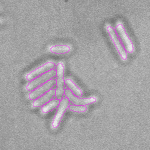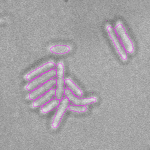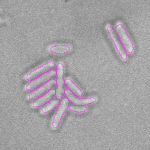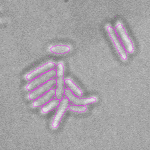
In Julou et al. 2013[1] the authors have shown that when cells form a single layer, a z-stack of transmitted-light images could be transformed into an even more contrasted images than a phase-contrast image (see figure below, left panel). Inspired by this result, I developed a neural-network that is able to take advantage of the information contained in the different slices of a defocused z-stack (typically 5-slices) of transmitted-light images to segment cells.
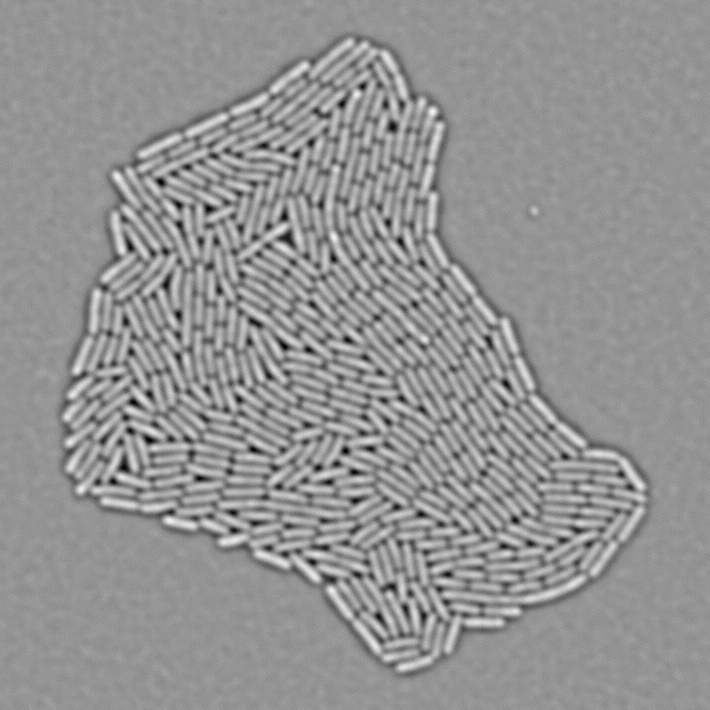
|
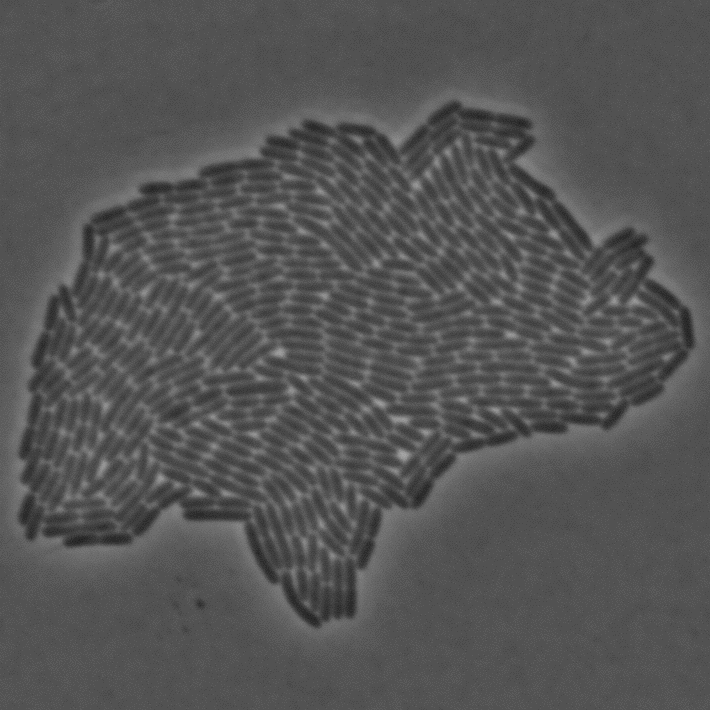
|
Method
Architecture
The architecture of the network is composed of two blocks:
- A block that transforms the 3D stack into a 2D multichannel images by applying a successively 3D convolutions and Z-maxpooling (maxpooling along Z-axis only). The resulting tensor has the same dimension along Y and X axis as the input image, no Z axis and several channels.
- A U-Net[2], a classical convolutional network with auto-encoder architecture involving skip connections, which can reproduce fine grained details while making use of higher-level spatially coarse information.
I compared the first block that makes use of the spatial information on the Z-axis with a block that contains only fully-connected layers along on the Z-axis and with the same number of parameter: I obtained much better results with the former one.
Segmentation with EDM
In order to segment bacteria, the network predicts a Euclidean Distance Map (see figure below), which is much more effective than predicting binary masks especially when cell density is high. The objective loss function is a L2 (a classical regression loss). I already used this technique in a previous work.

A classical watershed algorithm is then applied on the predicted EDM to segment cell instances. It requires an hyperparameter to decided wether two adjacent segmented regions are a single cell or not.
Data augmentation and processing
In order to limit the size of the necessary dataset and to improve generalization capacity of the network I applied the following data augmentation transforms:
- random normalization: images where normalized with the formula I’ = (I - C) / S with:
- C ∈ [ mean(I) ± f x std(I) ]
- S ∈ [ std(I) / f; std(I) x f ]
- and f a used-defined hyperparameter
- random Gaussian blurring
- random Gaussian noise addition
- elastic deformation
- random cropping
- random zooming with a random aspect ratio in a user-defined range
- random horizontal/vertical flipping and 90° rotation
To avoid unrealistic out-of-bounds artifacts, elastic deform was first performed in the full-size image, and zoom was performed during cropping.
Acknowledgments and Code Availability
- This study was a mission ordered by Prof. Meriem El Karoui. I especially thank Daniel Thédié whith whom I collaborated and all the lab for the feedbacks.
- Code is open source and available here.
- A wiki with an example dataset kindly provided by Daniel Thédié is available here
- Trained weights are available within BACMMAN software.
References
-
T. Julou et al., “Cell–cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies,” Proceedings of the National Academy of Sciences, vol. 110, no. 31, pp. 12577–12582, 2013.
-
O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, 2015, pp. 234–241, [Online]. Available at: https://arxiv.org/abs/1505.04597.
https://arxiv.org/abs/1505.04597
