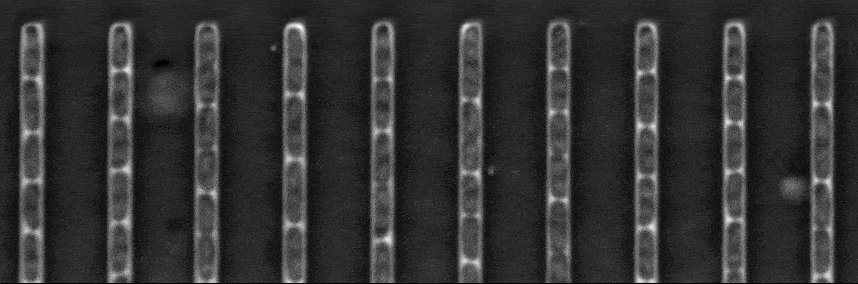
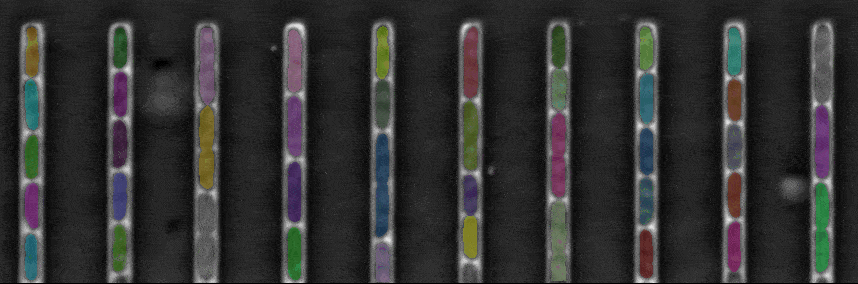
Segmentation in Transmitted-Light Images
21 December 2021
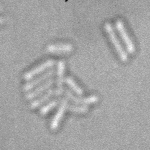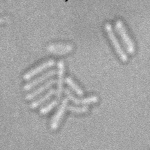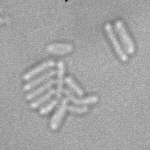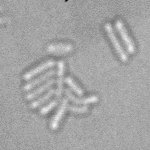
This is a novel cell segmentation method that tackles the difficult case in which neither a cell marker nor phase-contrast are available.
|

|
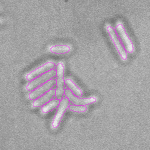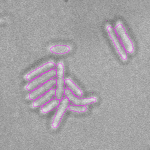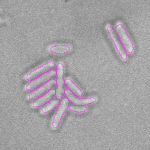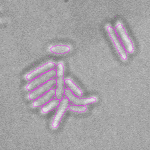
|
Self-Supervised Blind Denoising
12 June 2021
This post describes a novel state-of-the-art blind denoising method based on self-supervised deep neural networks[1] I am currently developing with Charles Ollion, Sylvain Le Corff (CMAP, Ecole Polytechnique, Université Paris-Saclay), Elisabeth Gassiat (Université Paris-Saclay, CNRS, Laboratoire de mathématiques d’Orsay) and Luc Lehéricy (Laboratoire J. A. Dieudonné, Université Côte d’Azur, CNRS). The preprint is available here: https://arxiv.org/abs/2102.08023


|


|
DistNet: Real-time Deep Detection of bacteria
19 March 2020
Along with Charles Ollion (CMAP, Ecole Polytechnique, Université Paris-Saclay), I recently developed DistNet[1], a method based on a novel Deep Neural Network (DNN) architecture that allows simultaneous segmentation and tracking of bacteria growing in the Mother Machine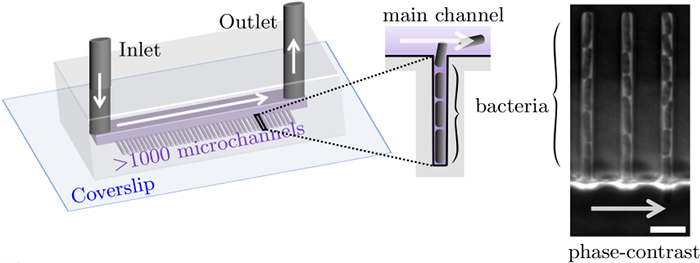
The Mother machine is a popular microfluidic device that allows long-term time-lapse imaging of thousands of cells in parallel by microscopy. It has become a valuable tool for single-cell level quantitative analysis and characterization of many cellular processes such as gene expression and regulation, mutagenesis or response to antibiotics.
White arrows represent the flow of growing medium in the mother machine microfluidic chip. Left: corresponding phase-contrast microscopy image showing Escherichia Coli bacteria growing in the microchannels. Scale bar: 5 μm.[2] microfluidic device at very low error rate. The article describing the method has been accepted in the MICCAI 2020 Conference.