
SABILab is a consulting company that aims at supporting scientific research by providing data/bio-image analysis expertise to scientists. It also has an academic research activity.
How Can We Help?
Can we assist you in your data analysis needs?
We offer a variety of services to help you improve your data production pipeline (data acquisition, image processing, statistical processing), including diagnostic evaluations, design and implementation of data analysis pipelines using open-source software, and tailored image processing methods. We also provide training in image analysis, deep learning, and statistical processing with R and Python. Check out our research works page for examples of our capabilities and let us know how we can assist you.
Jean Ollion
Founder of SABILab
 As a trained scientist with a double expertise in cell biology and bio-image analysis, I have extensive experience in both experimental work and data analysis.
During my PhD and post-doctoral studies, I developed two software tools: TANGO for 3D analysis of nuclear organization and BACMMAN for high-throughput analysis of Mother Machine
As a trained scientist with a double expertise in cell biology and bio-image analysis, I have extensive experience in both experimental work and data analysis.
During my PhD and post-doctoral studies, I developed two software tools: TANGO for 3D analysis of nuclear organization and BACMMAN for high-throughput analysis of Mother Machine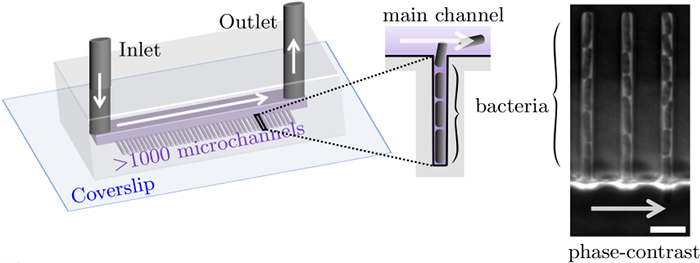
The Mother machine is a popular microfluidic device that allows long-term time-lapse imaging of thousands of cells in parallel by microscopy. It has become a valuable tool for single-cell level quantitative analysis and characterization of many cellular processes such as gene expression and regulation, mutagenesis or response to antibiotics. data.
I now work as a data scientist and bio-image analyst, providing expertise to other scientists. My background in biology allows me to understand the scientific and technical aspects of data analysis. I also continue to conduct research, recently developing an original bio-image processing method involving deep learning. I am currently working on a general purpose denoising method based on deep neural networks.
Education & Experience
- 2016 - 2017: Université Paris Saclay: Deep Learning course from Data Science Master’s degree (external candidate)
- 2015 - 2019: Laboratoire Jean Perrin CNRS / Sorbonne Université : Post-doctoral fellow: Mutation dynamics in single bacteria cells. Experience in microfluidics, microscopy, image processing and analysis
- 2010 - 2014: Dynamique et régulation des génomes, National Museum of Natural History: PhD Nuclear organization of centromeric DNA in human cells. Experience in cell biology, microscopy, image processing and analysis
- 2009 - 2010: Sorbonne Université: Master’s degree Molecular and cellular biology
- 2006 - 2010: Ecole polytechnique alumni. General scientific training - specialization in biology.
Publications
-
J. Ollion, C. Ollion, E. Gassiat, L. Lehéricy, and S. L. Corff, “Joint self-supervised blind denoising and noise estimation,” arXiv preprint arXiv:2102.08023, 2021.
https://arxiv.org/abs/2102.08023 -
J. Ollion and C. Ollion, “DistNet: Deep Tracking by displacement regression: application to bacteria growing in the Mother Machine,” 2020.
-
L. Robert, J. Ollion, and M. Elez, “Real-time visualization of mutations and their fitness effects in single bacteria,” Nature protocols, vol. 14, no. 11, pp. 3126–3143, 2019.
-
J. Ollion, M. Elez, and L. Robert, “High-throughput detection and tracking of cells and intracellular spots in mother machine experiments,” Nature protocols, vol. 14, no. 11, pp. 3144–3161, 2019, [Online]. Available at: https://rdcu.be/bRSze.
https://rdcu.be/bRSze -
L. Robert, J. Ollion, J. Robert, X. Song, I. Matic, and M. Elez, “Mutation dynamics and fitness effects followed in single cells,” Science, vol. 359, no. 6381, pp. 1283–1286, 2018, [Online]. Available at: https://science.sciencemag.org/content/359/6381/1283.editor-summary.
https://science.sciencemag.org/content/359/6381/1283.editor-summary -
J. Ollion, F. Loll, J. Cochennec, T. Boudier, and C. Escudé, “Proliferation-dependent positioning of individual centromeres in the interphase nucleus of human lymphoblastoid cell lines,” Molecular biology of the cell, vol. 26, no. 13, pp. 2550–2560, 2015, [Online]. Available at: https://www.molbiolcell.org/doi/full/10.1091/mbc.E14-05-1002.
https://www.molbiolcell.org/doi/full/10.1091/mbc.E14-05-1002 -
J. Ollion, J. Cochennec, F. Loll, C. Escudé, and T. Boudier, “Analysis of nuclear organization with TANGO, software for high-throughput quantitative analysis of 3D fluorescence microscopy images,” in The Nucleus, Springer, 2015, pp. 203–222.
https://link.springer.com/protocol/10.1007%2F978-1-4939-1680-1_16 -
J. Ollion, J. Cochennec, F. Loll, C. Escudé, and T. Boudier, “TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization,” Bioinformatics, vol. 29, no. 14, pp. 1840–1841, 2013, [Online]. Available at: https://academic.oup.com/bioinformatics/article/29/14/1840/231770.
https://academic.oup.com/bioinformatics/article/29/14/1840/231770
Contact
If you require any further information, feel free to contact me
