DistNet: Real-time Deep Detection of bacteria
19 March 2020
Along with Charles Ollion (CMAP, Ecole Polytechnique, Université Paris-Saclay), I recently developed DistNet[1], a method based on a novel Deep Neural Network (DNN) architecture that allows simultaneous segmentation and tracking of bacteria growing in the Mother Machine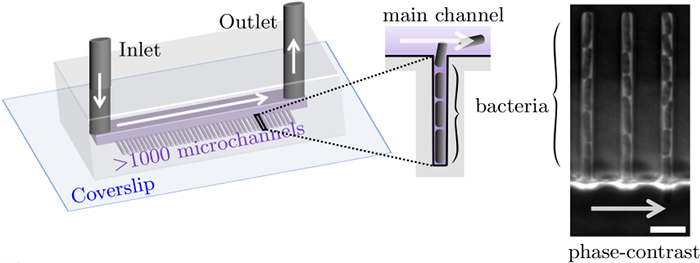
The Mother machine is a popular microfluidic device that allows long-term time-lapse imaging of thousands of cells in parallel by microscopy. It has become a valuable tool for single-cell level quantitative analysis and characterization of many cellular processes such as gene expression and regulation, mutagenesis or response to antibiotics.
White arrows represent the flow of growing medium in the mother machine microfluidic chip. Left: corresponding phase-contrast microscopy image showing Escherichia Coli bacteria growing in the microchannels. Scale bar: 5 μm.[2] microfluidic device at very low error rate. The article describing the method has been accepted in the MICCAI 2020 Conference.
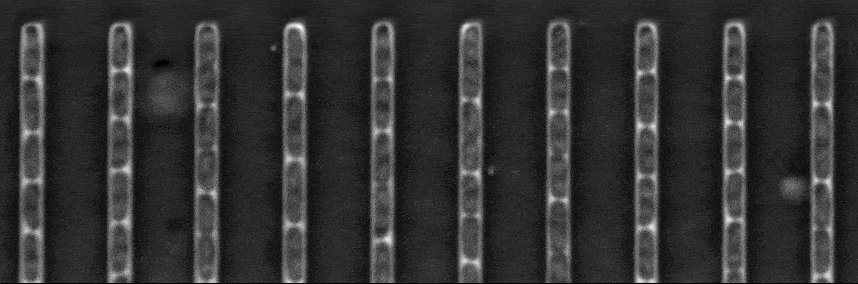
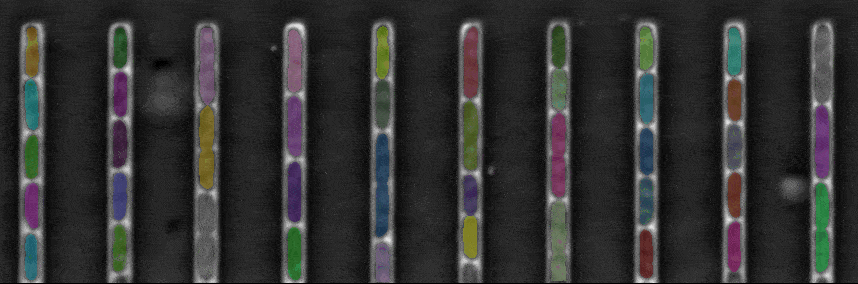
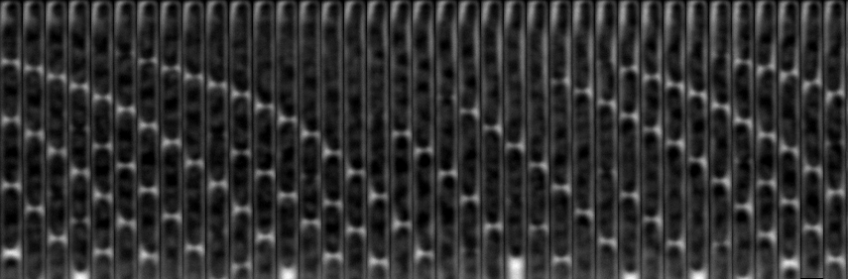
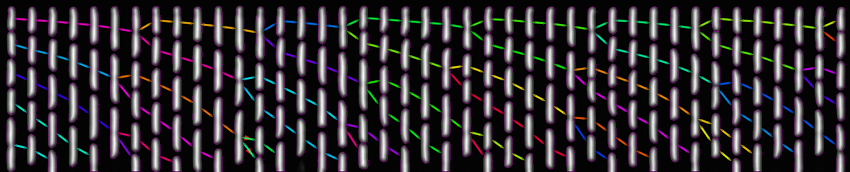
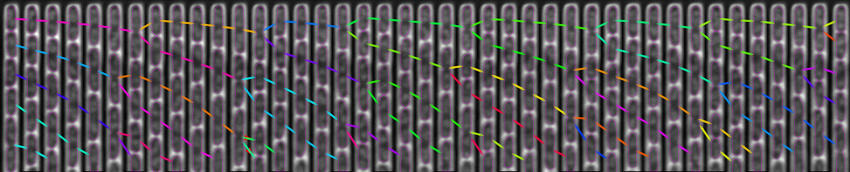
The figure above shows a phase-contrast movie of bacteria growing in the microchannels of the Mother Machine, as well as the result of the spatio-temporal detection (segmentation and tracking) made by DistNet. The next figure corresponds to another way of showing this data: successive frames of one single microchannel are displayed next to each other, referred to as kymograph. Result of segmentation is visible as cell outlines and result of tracking as coloured arrows.
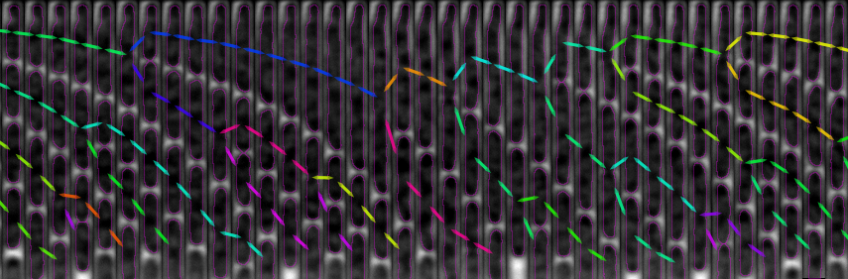

Time
Context
Tracking of bacteria growing in the mother machine faces three major challenges:
- Cell growth induces changes in bacteria morphology
- Bacteria can divide
- Due to cell growth, bacteria located at the open-end of microchannels are pushed out by other bacteria, thus their next observation is sometimes outside or partly outside the image.
Studying some biological processes such as mutagenesis requires very fine statistics in order to detect rare events as in [3]. To achieve this, one need to analyze massive datasets with typically 106-107 observations of bacteria, at a very low error rate in order to limit manual curation time.
Problem formulation
The main contribution of our approach is that we perform tracking by regression of the bacteria displacement between two successive frames. This allows to perform the tracking of multiple objects in one single prediction, with very common and simple network architectures (see next section). We also perform segmentation simultaneously by regression of the Euclidean Distance Map (EDM). We showed that performing segmentation and tracking simultaneously yielded in lower error rates compared to performing them separately.
This formulation contrasts with most current methods, that perform detection first, then tracking with one prediction per detected object.


Network architecture
Our network is based on U-Net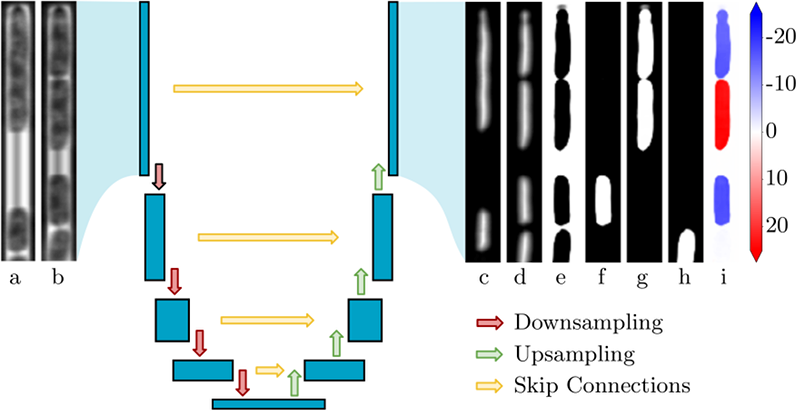
U-Net architecture.
Each blue block corresponds to a 2D multi-channel feature map (resulting from convolutions). The network has an encoder-decoder structure. The encoder reduces spatial dimensions and increases the number of channels at each contraction (red arrows). The decoder reduces the number of channels at each up-sampling level, and restores spatial dimensions using both feature maps of the previous level (green arrows) and of the corresponding level in the encoder (yellow arrows). Inputs are couples of successive grayscale images (a: previous frame, b: current frame). The upper bacteria in (a) divides in (b). Outputs are: EDM predictions for the previous (c) and current frame (d); category prediction (e): background, (f): cell that do not divide and are associated to a cell at the previous frame, (g): cells that divided, (h): cells that are not associated to a cell at the previous frame); (i): prediction of bacteria Y-displacement between the two frames, in pixels and within an image of height 256.[4], a widely-used network architecture in bio-image processing, that has the advantage of being simple and easy to train.
The originality of our method is to introduce a self-attention layer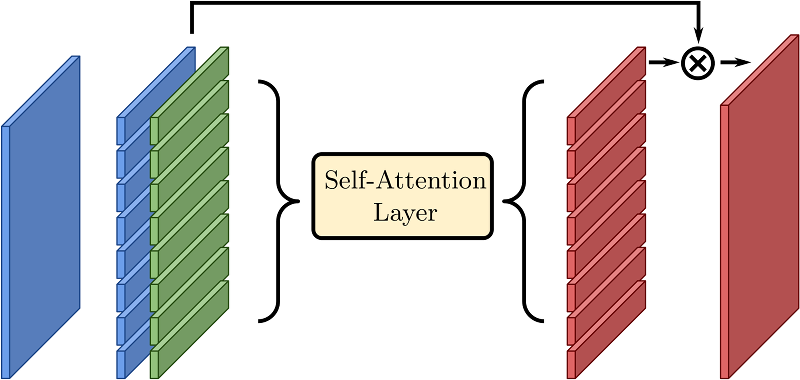
Self-attention layer.
A spatial feature map (left, blue) is interpreted as a set of feature vectors. These vectors are combined with positional embedding that only depend on their index (for instance i ∈ [0, 7] if the spatial dimensions are 8 × 1). The self-attention effectively transforms the set into a new one, where global information may be used. The final output has a skip connection with the input and can be re-interpreted as a spatial map.[5] in this network.
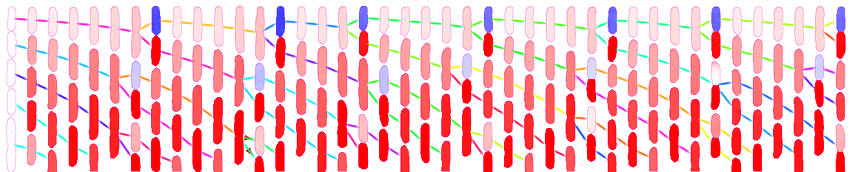
This layer enables the DNN to combine information from the whole image, while a convolution only mixes information locally. To illustrate this intuition, the next figure shows a few example of predictions and the associated attention weight matrix. A weight matrix can be read in the following way: for each output region (columns), it shows where the attention was mostly focused on the inputs (rows). For instance, a perfect diagonal attention matrix would mean that most of the information needed to produce an output region comes from the same input region.
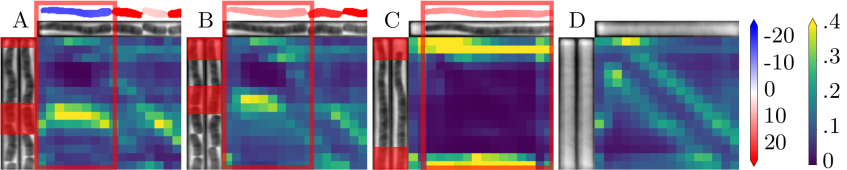
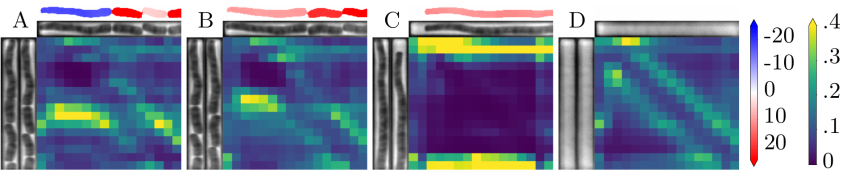
We observe here that in the case of long cells, self-attention focuses on the edges of cells rather than on their interior (panel C). This is particularly visible, in panel A where a division occurs, and the attention for upper daughter cell prediction is focused on the edges of the mother cell. Panel B corresponds to the next observation, and consistently, we observe that the attention moves upwards and is still focused on the edges of the previous cell.
Conclusion
This method presents several benefits:
- Global consistency: this method yields more coherent results, i.e. less conflicting predictions compared to a method that make one prediction per bacterium, because tracking is done simultaneously for all bacteria.
- Speed: This method is faster because one prediction by image is needed instead of one for the detection, then a second one per detected bacterium for the tracking.
- Simplicity / versatility: Our method enables to jointly train a single model, which is derived straightforwardly from a U-Net architecture, and could be adapted easily to different problem settings (e.g. 2D/3D geometries) or backbone networks. In contrast, tracking methods involving two steps and several models induce more hyper-parameters and complexity.
We applied successfully this method to the problem of bacteria growing in the mother machine, and achieved error rates inferior to 0.005% for tracking and of 0.03% for segmentation, outperforming current state-of-the-art methods, and making this method well-suited for high-throughput data analysis.
Implementation / Availability
An implementation of DistNet for tensorflow/keras is are available in this repository. In order to run training on cloud we developed an iterator that allows to read images in .h5 files, which is much faster on cloud than single image files.
To test DistNet, we provide a tutorial for a BACMMAN[6] module along with a sample dataset, as well as a tutorial to adapt it to other datasets with fine-tuning, on google colab, a service that provides free GPU.
References
-
J. Ollion and C. Ollion, “DistNet: Deep Tracking by displacement regression: application to bacteria growing in the Mother Machine,” 2020.
-
P. Wang et al., “Robust growth of Escherichia coli,” Current biology, vol. 20, no. 12, pp. 1099–1103, 2010.
-
L. Robert, J. Ollion, J. Robert, X. Song, I. Matic, and M. Elez, “Mutation dynamics and fitness effects followed in single cells,” Science, vol. 359, no. 6381, pp. 1283–1286, 2018, [Online]. Available at: https://science.sciencemag.org/content/359/6381/1283.editor-summary.
https://science.sciencemag.org/content/359/6381/1283.editor-summary -
O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, 2015, pp. 234–241, [Online]. Available at: https://arxiv.org/abs/1505.04597.
https://arxiv.org/abs/1505.04597 -
A. Vaswani et al., “Attention is all you need,” in Advances in neural information processing systems, 2017, pp. 5998–6008, [Online]. Available at: https://arxiv.org/abs/1706.03762.
https://arxiv.org/abs/1706.03762 -
J. Ollion, M. Elez, and L. Robert, “High-throughput detection and tracking of cells and intracellular spots in mother machine experiments,” Nature protocols, vol. 14, no. 11, pp. 3144–3161, 2019, [Online]. Available at: https://rdcu.be/bRSze.
https://rdcu.be/bRSze
